Stochastic motion and transcriptional dynamics of pairs of distal DNA loci on a compacted chromosome
David B. Brückner, Hongtao Chen, Lev Barinov, Benjamin Zoller, Thomas Gregor. Science 380, 1357-1362 (2023).
—
Abstract
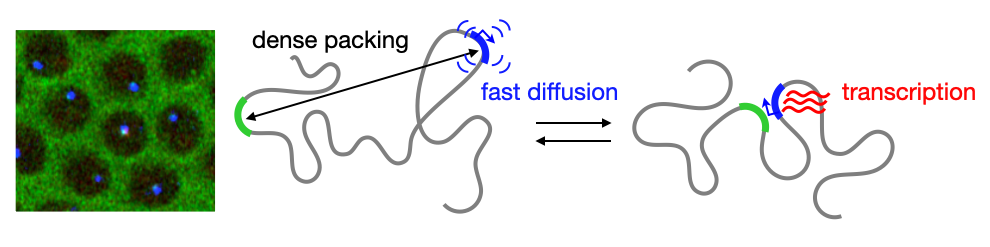
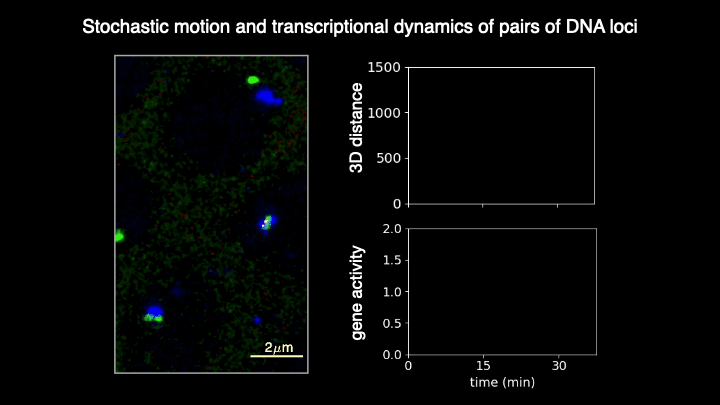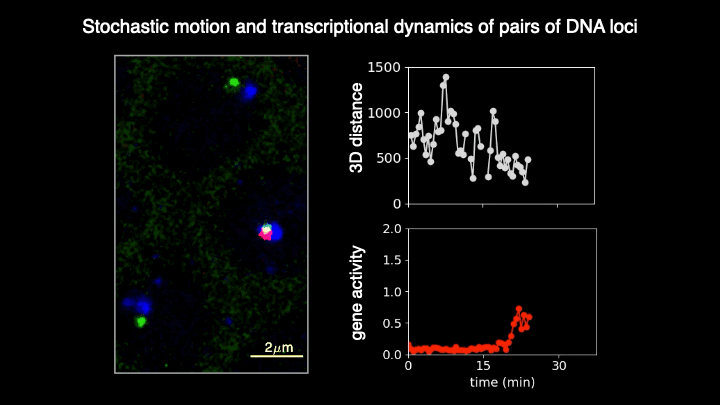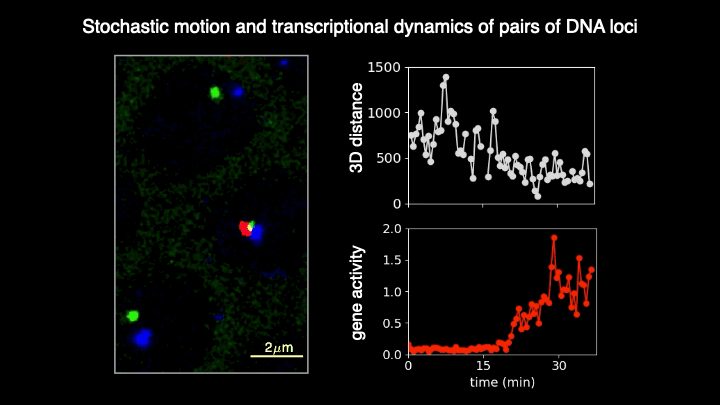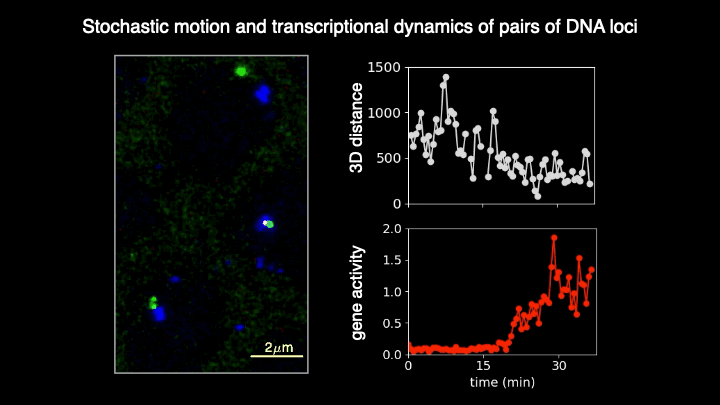
Chromosomes in the eukaryotic nucleus are highly compacted. However, for many functional processes, including transcription initiation, the pairwise motion of distal chromosomal elements such as enhancers and promoters is essential and necessitates dynamic fluidity. Here, we used a live-imaging assay to simultaneously measure the positions of pairs of enhancers and promoters and their transcriptional output while systematically varying the genomic separation between these two DNA loci. Our analysis reveals the coexistence of a compact globular organization and fast subdiffusive dynamics. These combined features cause an anomalous scaling of polymer relaxation times with genomic separation leading to long-ranged correlations. Thus, encounter times of DNA loci are much less dependent on genomic distance than predicted by existing polymer models, with potential consequences for eukaryotic gene expression.
Editor’s Summary
A crucial step in gene regulation is the physical encounter of dispersed enhancer-promoter pairs across the genome. However, how distal DNA elements find each other in the nuclear space remains unclear. Brückner et al. visualized the three-dimensional motion of pairs of DNA loci of varying separations along the chromosome and their transcriptional output in developing fly embryos. They found an unexpected combination of dense packing and rapid diffusion, leading to encounter times with a weak dependence on genomic separation. These results imply that transcriptional contacts are possible across large genomic distances, with crucial implications for gene regulation.
—Di Jiang (Science Magazine)