Optimal decoding of cellular identities in a genetic network
Mariela D. Petkova, Gasper Tkacik, William Bialek, Eric F. Wieschaus, and Thomas Gregor. Cell 176(4), 844–855 (2019).
Abstract
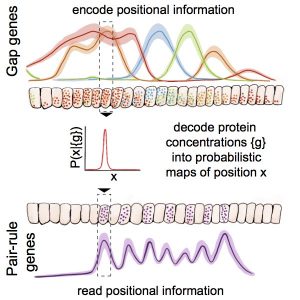 In developing organisms, spatially prescribed cell identities are thought to be determined by the expression levels of multiple genes. Quantitative tests of this idea, however, require a theoretical framework capable of exposing the rules and precision of cell specification over developmental time. Using the gap gene network in the early fly embryo as an example, we use such a framework to show how expression levels of the four gap genes can be jointly decoded into an optimal specification of position with 1% accuracy. The decoder correctly predicts, with no free parameters, the dynamics of pair-rule expression patterns at different developmental time points and in various mutant backgrounds. Precise cellular identities are thus available at the earliest stages of development, contrasting the prevailing view of positional information being slowly refined across successive layers of the patterning network. Our results suggest that developmental enhancers closely approximate a mathematically optimal decoding strategy.
In developing organisms, spatially prescribed cell identities are thought to be determined by the expression levels of multiple genes. Quantitative tests of this idea, however, require a theoretical framework capable of exposing the rules and precision of cell specification over developmental time. Using the gap gene network in the early fly embryo as an example, we use such a framework to show how expression levels of the four gap genes can be jointly decoded into an optimal specification of position with 1% accuracy. The decoder correctly predicts, with no free parameters, the dynamics of pair-rule expression patterns at different developmental time points and in various mutant backgrounds. Precise cellular identities are thus available at the earliest stages of development, contrasting the prevailing view of positional information being slowly refined across successive layers of the patterning network. Our results suggest that developmental enhancers closely approximate a mathematically optimal decoding strategy.
Quanta Magazine Article (PDF).
Cells optimize the use of information available to them to find their correct placement during fly embryo development. Information deposited in the embryo by the fly mother is translated into genetic and molecular instructions that drive the precise placement of stripes on the developing fly larva. Illustration by Mariela Petkova.