Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo
Jacques Bothma, Hernan Garcia, Samuel Ng, Michael W. Perry, Thomas Gregor, and Michael S. Levine. eLife 2015; 4:e07956 (2015).
Abstract
Metazoan genes are embedded in a rich milieu of regulatory information that often includes multiple enhancers possessing overlapping activities. In this study, we employ quantitative live imaging methods to assess the function of pairs of primary and shadow enhancers in the regulation of key patterning genes-knirps, hunchback, and snail-in developing Drosophila embryos. The knirps enhancers exhibit additive, sometimes even super-additive activities, consistent with classical gene fusion studies. In contrast, the hunchbackenhancers function sub-additively in anterior regions containing saturating levels of the Bicoid activator, but function additively in regions where there are diminishing levels of the Bicoid gradient. Strikingly sub-additive behavior is also observed for snail, whereby removal of the proximal enhancer causes a significant increase in gene expression. Quantitative modeling of enhancer–promoter interactions suggests that weakly active enhancers function additively while strong enhancers behave sub-additively due to competition with the target promoter.
PDF.
eLife digest
 Only a subset of the genes in a cell is active at any time. Gene activation or ‘transcription’ is controlled by specific DNA sequences called promoters and enhancers. Promoters are found next to genes and recruit the protein machinery needed to transcribe the gene. Enhancers are located further away from genes and interact with the promoter to increase gene transcription.
Only a subset of the genes in a cell is active at any time. Gene activation or ‘transcription’ is controlled by specific DNA sequences called promoters and enhancers. Promoters are found next to genes and recruit the protein machinery needed to transcribe the gene. Enhancers are located further away from genes and interact with the promoter to increase gene transcription.
Some genes have multiple enhancers with overlapping activities, typically including a primary enhancer and secondary or ‘shadow’ enhancer that is even more distant. Multiple enhancers are used to control gene transcription in different types of cells. For example, shortly after fertilization around 1000 enhancers in the embryo of the fruit fly Drosophila regulate several hundred genes that are important for development. These activities establish which cell type any given cell in the embryo will become. Many of these developmental genes have shadow enhancers, which are thought to make gene activity more precise and reliable. It is not known, however, how several enhancers interact with the same promoter at the same time. For example, do the enhancers’ individual effects add together? Or can the combined effects of multiple enhancers be more (or even less) than the sum of their parts?
Bothma, Garcia et al. have now examined how multiple enhancers regulate the activity of three developmental genes (called hunchback, knirps, andsnail) in early Drosophila embryos. The experiments showed that the individual effects of the knirps enhancers add together as one might expect. On the other hand, the snail enhancers interfere with each other, which means that their combined effect on transcription is less than the sum of the two individual effects. Furthermore, other experiments revealed that the combined effect of the hunchback enhancers depends upon whether another component that is needed for gene activation is in short supply.
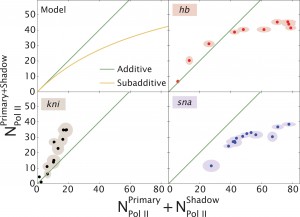 To understand these observations, Bothma, Garcia et al. then developed a mathematical model. The model proposes that behavior of enhancers depends upon how strongly they interact with the target promoter. Since ‘weak’ enhancers do not interact very often, their effects can easily add together. However, the effects of ‘strong’ enhancers do not add together because they often compete to interact with the promoter. These findings show how multiple enhancers can work in a complex manner to control gene transcription.
To understand these observations, Bothma, Garcia et al. then developed a mathematical model. The model proposes that behavior of enhancers depends upon how strongly they interact with the target promoter. Since ‘weak’ enhancers do not interact very often, their effects can easily add together. However, the effects of ‘strong’ enhancers do not add together because they often compete to interact with the promoter. These findings show how multiple enhancers can work in a complex manner to control gene transcription.